
Report from a BOU-funded project
In 2008 a new storm petrel species was described (Bolton et al. 2008), endemic to The Azores archipelago: Monteiro’s Storm Petrel (Hydrobates monteiroi) breeds during the summer months, a sympatric sister species to the more widely distributed Band-rumped Storm Petrel (Hydrobates castro), which also breeds in the Azores, but during the winter months. These two seasonally distinct storm petrels were initially thought to belong to the same species, but with two seasonally distinct breeding populations (Monteiro et al. 1996). Comparisons of morphometrics, burrowing calls, diet and mitochondrial DNA led to these two breeding populations to be recognised as two separate, sympatric species (Bolton et al. 2008; Friesen et al. 2007; Smith et al. 2007), further reinforced by subsequent work (Silva et al. 2016 and Taylor et al. 2019). Additional nuclear characterisation of both species would however be useful to further illuminate their speciation history, especially with focus on particular genes of interest that are prone to the pressure of selection. One such example of genes that are the target of selection are those that code for the Major Histocompatibility Complex (MHC). This MHC region was a main focus of the author’s PhD studies at Cardiff University.

Figure 1 Monteiro’s Storm Petrel (Hydrobates monteiroi) chick © Alexandra McCubbin.
The MHC plays a key role in the adaptive immune response, coding for highly specific cell-surface proteins that fight off pathogens. The MHC is polygenic resulting in individuals with highly specific and varied MHC profiles (Janeaway et al. 2001). Notably, increased diversity tends to convey the ability to fight a wider range of pathogens (Penn et al. 2002). This fitness advantage can be selectively maintained, with MHC genes becoming targets for disease-based or sexual selection. Disease-based selection implies MHC genes have co-evolved with a variety of pathogens (Bernatchez and Landry 2003). Where MHC profiles are targets for sexual selection, MHC composition may also influence the mate choices of individuals, to better equip offspring with a fitness advantage. MHC-based mate choice has been described across many different animal taxa, with targets for mate choice varying – individuals may choose mates with specific MHC genes, or they may choose mates that result in a greater level of heterozygosity in offspring. To make such decisions, individuals must be able to detect MHC profile. It is thought this is achieved through scent, making species systems with well-developed olfactory capabilities interesting as case-studies for MHC expression and evolution (Boehm and Zufall 2006). One such group are the Procellariiform order of seabirds, which have a large, well-developed olfactory bulb (Bang 1966), and includes storm petrels such as H. castro and H. monteiroi.

Figure 2 Praia islet in the Azores, one of the islets that supports the breeding of the endemic H. monteiroi in the summer months, as well as the breeding of the widely-distributed H. castro during the winter months.
Previous research into avian MHC has shown that an ancient duplication of the MHC occurred, resulting in two lineages – DAB1 and DAB2 (Burri et al. 2008, Goebel et al. 2017). This was most likely prior to the radiation of all extant birds, with phylogenetic analysis showing that MHC sequences cluster by DAB locus and not by species. Research suggests that the MHC evolves through a ‘birth and death model’, where duplication events result in the creation of gene copies, which in turn can be lost from some genomes through deletion events (Burri et al. 2008, Wang et al. 2013, Dearborn et al. 2015). Previous MHC research concerning H. castro and H. monteiroi has been limited, with preliminary estimates of allele numbers provided, but prior to this PhD thesis there was no evidence of the second DAB lineage being present in the two species (Burri et al. 2014).
Classifying MHC in H. castro and H. monteiroi
DNA was first extracted from blood samples of both species, and amplification of the MHC gene region was then carried out using Polymerase Chain Reaction (PCR). Using an alignment of 11 previously published sequences on GenBank (five from H. castro and six from H. monteiroi), two sets of overlapping primers were developed to amplify a large region of the MHC DAB1 lineage for both species. Eleven primers were designed and used in seven pairwise combinations for testing in PCR. Following PCR optimisation, PCR products were sent for Sanger sequencing. Four primer combinations yielded clear results for the MHC Class IIB region in the DAB1 lineage. When attempting to amplify a large fragment for DAB2, it was found that previously published DAB2 primers from the outer region in Dearborn et al. (2015) worked to amplify H. castro and H. monteiroi, despite being developed for Leach’s Storm Petrel (Oceanodroma leucorhoa).
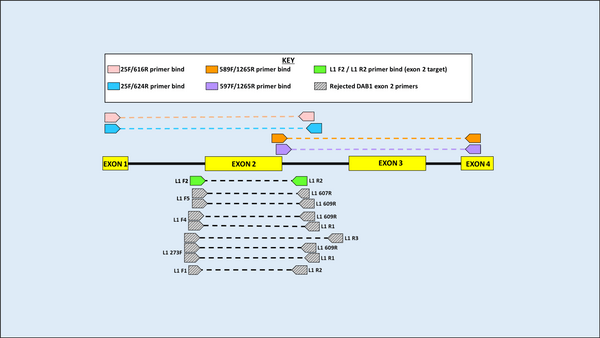
Figure 3 Primer binding and testing sites for the DAB1 lineage. Primers above the amplicon fragment show those developed in initial attempts to amplify the whole DAB1 fragment, in two overlapping segments. Below, grey represents primers used in exploratory work that did not pass quality checks. Green are primers ultimately used to amplify exon 2 and will be used in further work (names are not yet finalised).
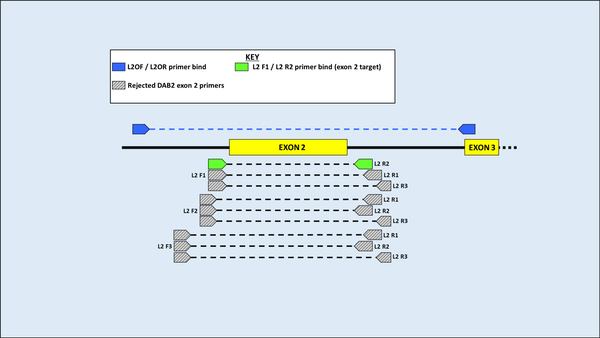
Figure 4 Primer binding and testing sites for the DAB2 lineage. Primers above the amplicon are those from Dearborn et al. (2015) that were initially used to amplify a large DAB2 fragment. Below, grey primers are those used in exploratory work that did not pass quality checks for sequencing. Green are the primers ultimately used to amplify exon 2 and will be used in further work (names are not yet finalised).
Once large fragments for both DAB lineages had been amplified, resulting sequences were used to design new primers specifically targeting exon 2 of the MHC Class IIB fragment, as the exon coding for the protein-binding site of the MHC and therefore a more important target for selection. These primers were designed by eye for both DAB lineages, purposefully choosing heterozygous sites around the exon that may indicate that >1 MHC copy is present. A total of 13 and nine primer combinations were trialled for DAB1 and DAB2 respectively. PCR optimisation and Sanger sequencing resulted in the successful development of a primer pair for amplifying each DAB lineage, in a way that captured heterozygous sequences, indicative that >1 MHC copy being amplified in individuals.
This was the first such amplification of two DAB lineages in H. castro and H. monteiroi and presented the novel discovery of a DAB2 lineage. This was confirmed by creating a phylogenetic tree using these sequences with other confirmed DAB1 and DAB2 sequences from other species. The sequences generated here separated into two distinct clades (or groups), representative of two DAB lineages rather than separating into different species groups. H. castro and H. monteiroi sequences generated using the DAB1 primers separated into a clade with other known DAB1 sequences, with the same pattern observed for sequences generated from DAB2 primers. This separation of DAB sequences demonstrates that the ancient duplication of the two lineages is retained in H.castro and H. monteiroi.
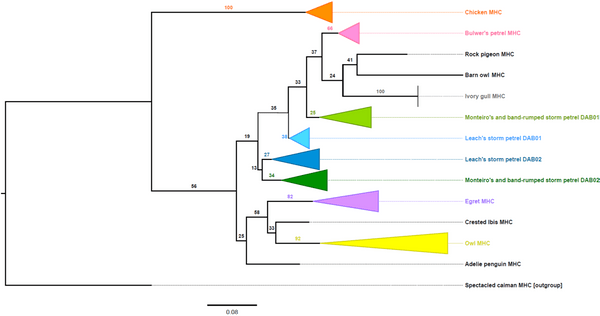
Figure 5 Maximum likelihood phylogenetic tree of MHC Class IIB exon 2 sequences in multiple bird species, created using IQTree (Nguyen et al. 2015). Numbers on branches show bootstrap support. The tree contains class IIB sequences from both DAB lineages in some species. The clade arrangements show that both DAB lineages are present in H. castro and H. monteiroi, and have been successfully amplified using this new method. An asterisk (*) specifies where MHC DAB lineage was unknown.
What’s next?
The primers developed here allow for further analysis of variability in this region and will contribute to future genetic comparisons and subsequent understanding of the two species. Through specialised sequencing techniques, the complex MHC profiles of individual birds can be characterised and compared both within and between species, contributing to further research on these two species. Comprehensive characterisation of the MHC is important for understanding the complex interplay of MHC, selection, mate choice and survival, and cold prove useful in future conservation efforts for the vulnerable H. monteiroi. The development of these primers demonstrates the first steps in achieving this characterisation, opening many exciting avenues for future research.
Funding
Alexandra McCubbin (PhD Candidate, Cardiff University, UK) was awarded an ornithological research grant of £1,500 in 2017 for a project entitled ‘Interaction of genetic isolation and mate choice in a recently evolved species pair of Atlantic Storm-Petrels’.
References
Bang, B.G. 1966. The olfactory apparatus of tubenosed birds (Procellariiformes). Acta Anat 65: 391-415. VIEW
Bernatchez, L. & Landry, C. 2003. MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years? Journal of Evolutionary Biology 16: 363-377. VIEW
Boehm, T. & Zufall, F. 2006. MHC peptides and the sensory evaluation of genotype. Trends in Neurosciences 29: 100-107. VIEW
Bolton, M. et al. 2008. Monteiro’s Storm-petrel Oceanodroma monteiroi: A new species from the Azores. Ibis 150: 717-727. VIEW
Burri, R., Hirzel, H.N., Salamin, N., Roulin, A. & Fumagalli, L. 2008. Evolutionary patterns of MHC class II B in owls and their implications for the understanding of avian MHC evolution. Molecular Biology and Evolution 25: 1180-1191. VIEW
Burri, R., Promerová, M., Goebel, J. and Fumagalli, L. 2014. PCR-based isolation of multigene families: Lessons from the avian MHC class IIB. Molecular Ecology Resources 14: 778-788. VIEW
Dearborn, D.C., Gager, A.B., Gilmour, M.E., McArthur, A.G., Hinerfeld, D.A. & Mauck, R.A. 2015. Non-neutral evolution and reciprocal monophyly of two expressed Mhc class II B genes in Leach’s storm-petrel. Immunogenetics 67: 111-123. VIEW
Friesen, V.L., Smith, a L., Gómez-Díaz, E., Bolton, M., Furness, R.W., González-Solís, J. & Monteiro, L.R. 2007. Sympatric speciation by allochrony in a seabird. Proceedings of the National Academy of Sciences of the United States of America 104: 18589-18594. VIEW
Goebel, J. et al. 2017. 100 million years of multigene family evolution: origin and evolution of the avian MHC class IIB. BMC Genomics 18: 1-9. VIEW
Janeaway, C.J., Travers, P., Walport, M. & Shlomchik, M. 2001. The two classes of MHC molecule are expressed differentially on cells. In: Immunobiology: The Immune System in Health and Disease. 5th edition. New York: Garland Science.
Monteiro, L.R., Ramos, R.A. & Furness, R.W. 1996. Past and present status and conservation of the seabirds breeding in the Azores archipelago. Biological Conservation 78: 319-328. VIEW
Penn, D.J., Damjanovich, K. & Potts, W.K. 2002. MHC heterozygosity confers a selective advantage against multiple-strain infections. Proceedings of the National Academy of Sciences 99: 11260-11264. VIEW
Silva, M.F., Smith, A.L., Friesen, V.L., Bried, J., Hasegawa, O., Coelho, M.M. & Silva, M.C. 2016. Mechanisms of global diversification in the marine species Madeiran Storm-petrel Oceanodroma castro and Monteiro’s Storm-petrel O. monteiroi: insights from a multi-locus approach. Molecular Phylogenetics and Evolution 98: 314-323. VIEW
Smith, A.L., Monteiro, L., Hasegawa, O. & Friesen, V.L. 2007. Global phylogeography of the band-rumped storm-petrel (Oceanodroma castro; Procellariiformes: Hydrobatidae). Molecular Phylogenetics and Evolution 43: 755-773. VIEW
Taylor, R.S. et al. 2019. Cryptic species and independent origins of allochronic populations within a seabird species complex (Hydrobates spp.). Molecular Phylogenetics and Evolution 139: 106552. VIEW
Wang, Z., Zhou, X., Lin, Q., Fang, W. & Chen, X. 2013. Characterization, Polymorphism and Selection of Major Histocompatibility Complex (MHC) DAB Genes in Vulnerable Chinese Egret (Egretta eulophotes). PLoS ONE 8: e74185. VIEW
Image credit
Top right: Band-rumped Petrel From The Crossley ID Guide Eastern Birds © Richard Crossley.
If you want to write about your research in #theBOUblog, then please see here.




