LINKED BOU SMALL RESEARCH GRANT
Searching for the genomic basis behind phenotypic divergence with low levels of genetic differentiation in the polytypic Rufous-collared Sparrow (Zonotrichia capensis) using whole-genome sequencing.
High-throughput sequencing is allowing researchers to unveil the genomic basis and processes behind avian plumage color variation (Funk & Taylor, 2019). Species with low background genetic differentiation between populations that vary in a certain plumage trait are particularly interesting, since they provide the opportunity to look for highly divergent genomic areas that might be linked to differences in coloration (Campagna et al., 2017; Funk & Taylor, 2019). The Rufous-collared Sparrow (Zonotrichia capensis) is a widespread Neotropical passerine that shows phenotypic and behavioral variation throughout its continental distribution from Mexico to Argentina, being one of the most polytipic birds in the world with 25 subspecies (Handford, 1983; Rising & Jaramillo, 2020). However, many of these subspecies are poorly differentiated in phenotype and geographical variation in morphology and behavior does not match subspecific taxonomy (Nottebohm, 1969; Handford, 1985). One of the few exceptions to this overall pattern is Z. c. australis (Fig. 1), the southernmost subspecies found in austral Chile and Argentina (Patagonia), which differentiates from all other subspecies by lacking the characteristic black lateral crownstripes (i.e., head uniformly grey or with only subtle traces of black; Rising & Jaramillo, 2020). Three mitochondrial lineages exist throughout the species’ distribution, and two of them occur in Argentina with signs of extensive gene flow (Lougheed et al., 2013; Campagna et al., 2014). Even though mitochondrial lineages do not match subspecies’ boundaries in general, Lougheed et al. (2013) recovered Z. c. australis as a slightly divergent (~ 0.7%) subclade within the most widespread lineage of the species in Argentina.
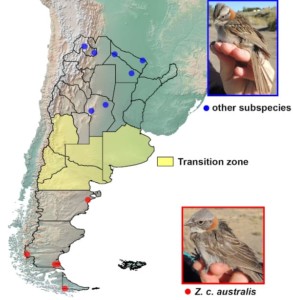
Figure 1 Geographic precedence of individuals representing Z. c. australis (grey head) and other subspecies (black crownstripes) depicted over an elevation map of southern South America with Argentinian provinces delimited. The transition zone, which was intentionally avoided for this first study since individuals show intermediate phenotypes, appears in yellow
In this context, we used whole-genome resequencing to investigate the genomic basis behind phenotypic differentiation between Z. c. australis and other subspecies from the southern region of its distribution, which seems to have evolved relatively rapidly and with low levels of genetic divergence. We obtained whole genomes of nine individuals from Patagonia representing Z. c. australis, and nine individuals from northcentral Argentina that possess the black crownstripes (Fig. 1). Since the mitochondrial lineage comprises at least eight subspecies including Z. c. australis, all individuals not representative of the latter were grouped and analyzed together for simplicity. For this first assessment, we have included individuals that represent the two extremes of plumage color variation within Z. capensis in Argentina (Fig.1), intentionally avoiding a contact zone where individuals show intermediate phenotypes, most likely as a result of gene flow between Z. c. australis and other subspecies. After processing the genomic data, we retained ~11 million single nucleotide polymorphism (SNPs) that were used for downstream analyses. Nuclear data was complemented with mitochondrial genomes, which were assembled from raw sequencing reads from the same individuals.
The mitochondrial gene tree confirmed that all individuals sampled belong to the same lineage, and showed high genetic variation within Z. c. australis, which was not recovered as a monophyletic lineage (Fig. 2). On the other hand, all other individuals from northern Argentina clustered together with high support and showed remarkably lower genetic diversity (Fig. 2). A principal component analysis (PCA) derived from the analysis of ~11 million nuclear SNPs showed a shallow but relatively clear separation between the two groups (Fig. 2). Contrary to what was observed at the mitochondrial DNA, nuclear genomic variation was lower within Z. c. australis (Fig. 2). The assessment of genetic population structure evidenced that the most likely number of genetic clusters four our sample is one (Fig. 2), in agreement with previous results from DNA microsatellite data that showed no phylogeographic structure within Argentina (Campagna et al., 2014). Concordantly, mean genomic differentiation between Z. c. australis and all other individuals based on ~ 11 million SNPs was low (FST = 0.015 ± 0.018; mean ± standard deviation, SD). However, we were able to find three divergence peaks on scaffolds 42, 81 and 653 that contrast with this background of low genetic differentiation (Fig. 3). Genomic differentiation between the two groups at these three peaks alone (1,350 SNPs) was remarkably higher (FST = 0.29 ± 0.09) than that calculated for the entire dataset (Fig. 3). Out of a total of 235 SNPs with an FST ≥ 0.8 found in these three regions combined, 211 were located within scaffold 42, which was the widest (~200 kb), most divergent (mean FST = 0.31 ± 0.39) genomic region identified (Fig. 4). Moreover, of a total of 53 SNPs fixed (FST = 1) between the two groups, 51 were located within scaffold 42. The other two peaks corresponded to two non-consecutive 15-kb windows within scaffold 81 (mean FST = 0.21 ± 0.29; 22 SNPs with FST ≥ 0.8 and 1 fixed SNP; Fig. 3), and one 15-kb window from scaffold 653 (mean FST = 0.22 ± 0.25; 2 SNPs with FST ≥ 0.8 and 1 fixed SNP; Fig. 3). Using the annotated genomes of the White-throated Sparrow (Z. albicollis) and the Zebra Finch (Taeniopygia guttata) we were able to map the scaffold 42 to chromosome 5 and observed that outlier SNPs were concentrated within and downstream the Suppression of tumorigenicity 5 (ST5) gene (Fig. 4). The two windows from scaffold 81 were mapped to the mitogen-activated protein kinase 3 (MAP4K3) gene and to a region downstream of the mitochondrial ribosomal protein S5 (MRPS5) on chromosome 3, while the peak from scaffold 653 was located within the solute carrier organic anion transporter family member 4C1 (SLC04C1) gene on the Z chromosome.
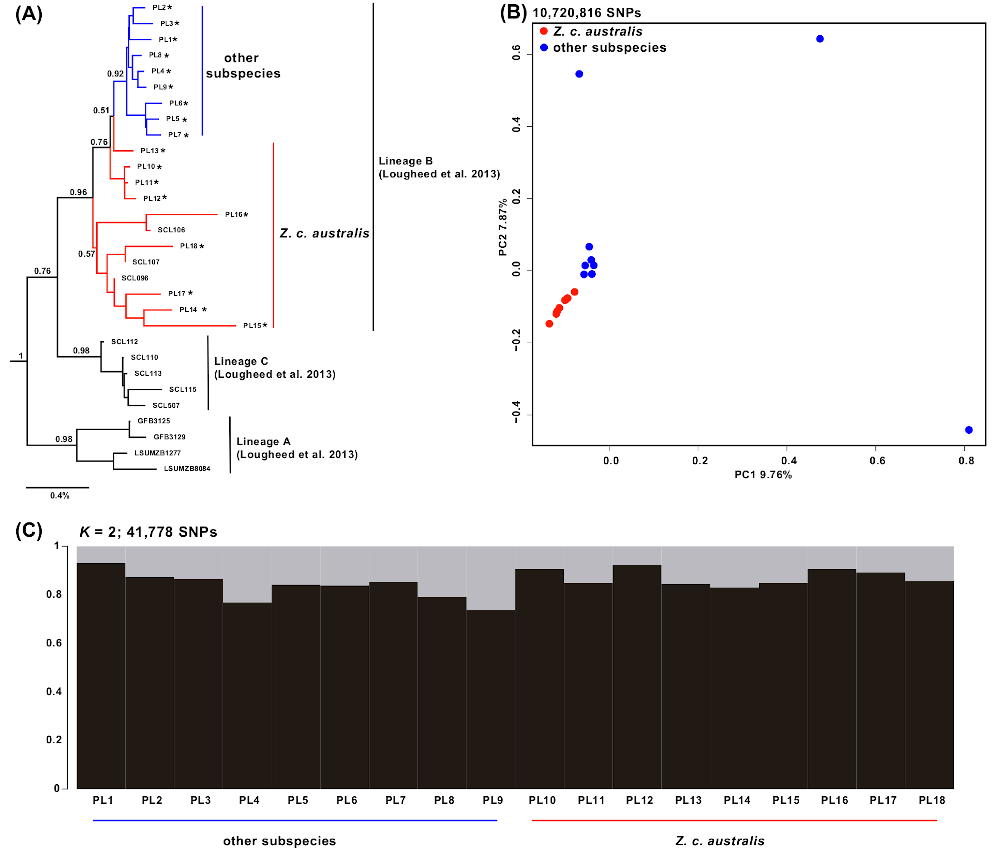 Figure 2 Mitochondrial and nuclear genomic differentiation between Z. c. australis and other subspecies. (A) Neighbor-joining tree constructed from four mitochondrial markers (COI, ND2, 16S and control region), showing the three lineages that exist within Z. capensis throughout its distribution (Lougheed et al. 2013). Numbers above or below the branches indicate node support based on 500 bootstrap pseudoreplicates. The 18 individuals whose whole genomes were sequenced for this project (PL1–PL18, lineage B) are denoted by an asterisk (all other sequences were obtained from Lougheed et al., 2013). (B) Principal component analysis based on ~ 11 million SNPs. (C) STRUCTURE plot for K = 2 based on 41,778 SNPs (excluding variants within 15 kb of each other in order to avoid linkage disequilibrium) showing that there are no genetic clusters distinguishable within Z. capensis. Click to view larger
Figure 2 Mitochondrial and nuclear genomic differentiation between Z. c. australis and other subspecies. (A) Neighbor-joining tree constructed from four mitochondrial markers (COI, ND2, 16S and control region), showing the three lineages that exist within Z. capensis throughout its distribution (Lougheed et al. 2013). Numbers above or below the branches indicate node support based on 500 bootstrap pseudoreplicates. The 18 individuals whose whole genomes were sequenced for this project (PL1–PL18, lineage B) are denoted by an asterisk (all other sequences were obtained from Lougheed et al., 2013). (B) Principal component analysis based on ~ 11 million SNPs. (C) STRUCTURE plot for K = 2 based on 41,778 SNPs (excluding variants within 15 kb of each other in order to avoid linkage disequilibrium) showing that there are no genetic clusters distinguishable within Z. capensis. Click to view larger
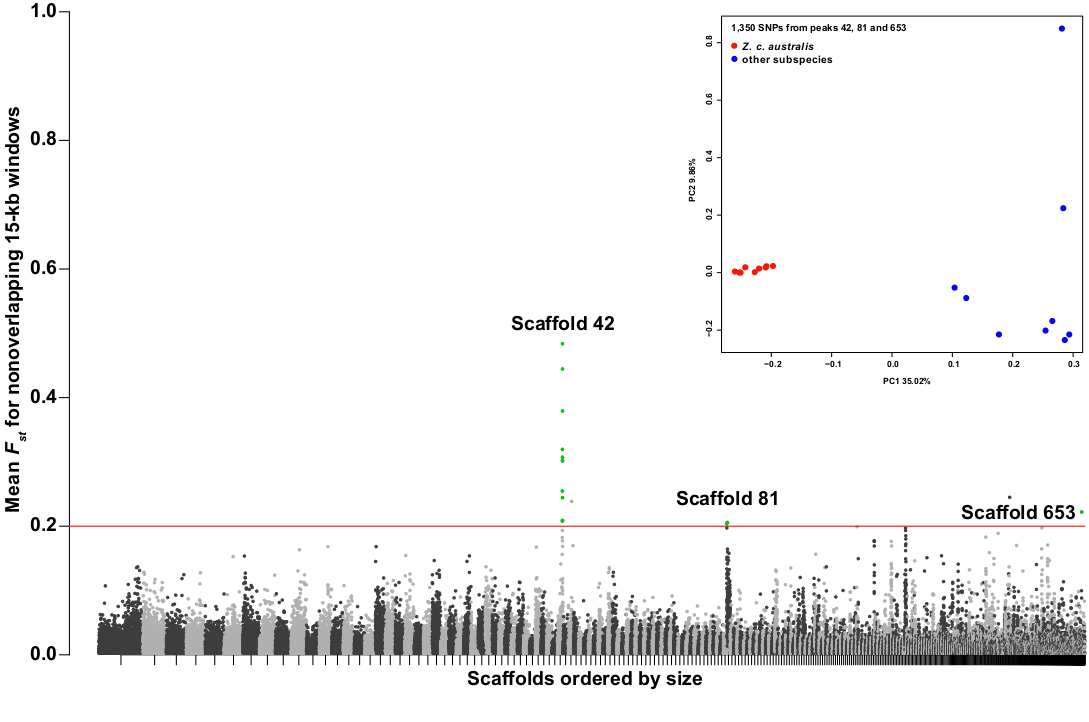 Figure 3 Manhattan plot for the comparison between Z. c. australis and all other individuals from northern Argentina. Each circle corresponds to the mean FST between groups for all the SNPs found within a nonoverlapping 15-kb window. Scaffolds are sorted by decreasing size and indicated by alternating black and gray colors. The red line at FST = 0.2 indicates the threshold used for the identification of divergent windows, which corresponds to regions that fall more than 10 SDs above the mean FST across all windows (FST = 0.015 ± 0.018). Dots highlighted in green are the 15-kb windows identified within three main divergence peaks on the scaffolds 42, 81 and 653. The inset on the upper right shows the PCA derived from the analysis of 1,350 SNPs under these three peaks. Click to view larger
Figure 3 Manhattan plot for the comparison between Z. c. australis and all other individuals from northern Argentina. Each circle corresponds to the mean FST between groups for all the SNPs found within a nonoverlapping 15-kb window. Scaffolds are sorted by decreasing size and indicated by alternating black and gray colors. The red line at FST = 0.2 indicates the threshold used for the identification of divergent windows, which corresponds to regions that fall more than 10 SDs above the mean FST across all windows (FST = 0.015 ± 0.018). Dots highlighted in green are the 15-kb windows identified within three main divergence peaks on the scaffolds 42, 81 and 653. The inset on the upper right shows the PCA derived from the analysis of 1,350 SNPs under these three peaks. Click to view larger
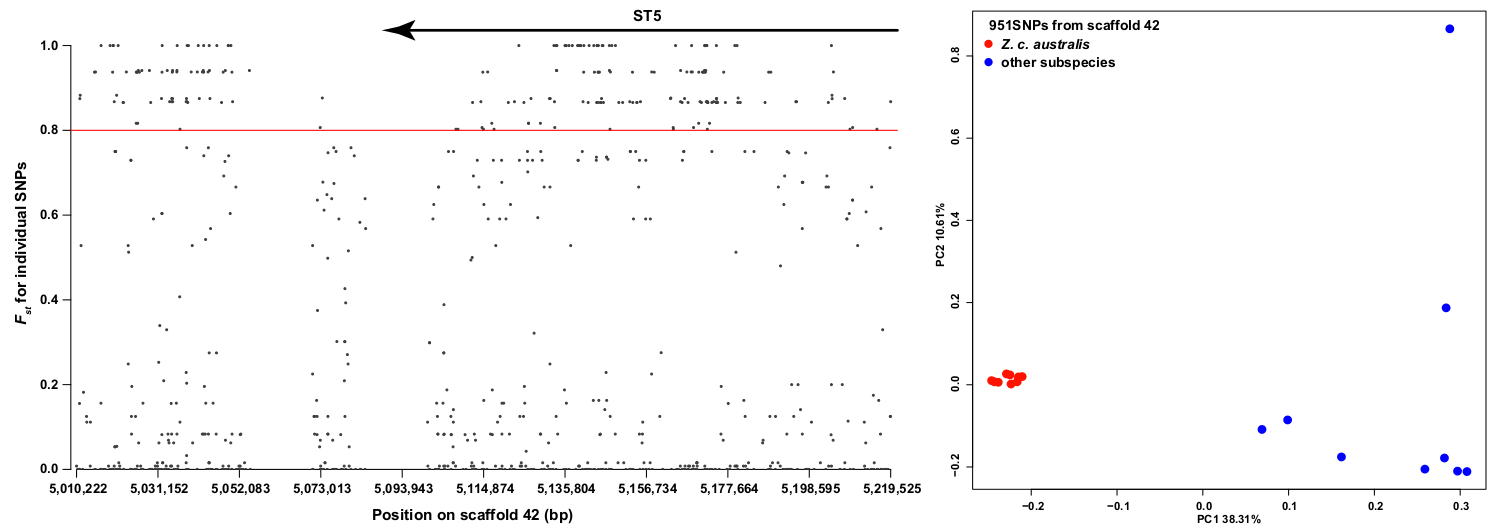 Figure 4 Individual FST and genomic position for all the SNPs found within the divergence peak that corresponds to scaffold 42. From 951 total SNPs, 211 have and FST ≥ 0.8 (red line) and 51 are fixed (FST = 1) between Z. c. australis and all other individuals. The position of the ST5 gene is indicated above by the arrow. Right: PCA derived from the analysis of the 951 SNPs from the divergence peak. Click to view larger
Figure 4 Individual FST and genomic position for all the SNPs found within the divergence peak that corresponds to scaffold 42. From 951 total SNPs, 211 have and FST ≥ 0.8 (red line) and 51 are fixed (FST = 1) between Z. c. australis and all other individuals. The position of the ST5 gene is indicated above by the arrow. Right: PCA derived from the analysis of the 951 SNPs from the divergence peak. Click to view larger
Even though we cannot discard the possibility that multiple genes are responsible for the phenotypic differences between Z. c. australis and other subspecies, our results point out to one particular genomic region that could constitute a new candidate gene for plumage color differentiation. This ~200-kb divergence peak, which concentrates 90% of the outlier SNPs and virtually all fixed differences between the two groups, is located within and downstream the ST5 gene, which seems to be involved in cytoskeletal organization and tumorigenicity in humans (Majidi et al., 1998), but has no characterized function in birds. Our findings add to the small but growing list of cases where a genomic region with no previous association with pigmentation is proposed as a candidate gene for plumage color differentiation (Küpper et al., 2015; Tuttle et al., 2016; Vickrey et al., 2018; Funk & Taylor, 2019). While further analyses are still needed, this study provides new, valuable information to advance our understanding of the genomic basis of plumage color variation in birds in general and within Z. capensis in particular.
I thank my collaborators and co-authors Leonardo Campagna from the Cornell Lab of Ornithology and Ana S. Barreira, Pablo L. Tubaro and Darío A. Lijtmaer from the Ornithology Division of the MACN-CONICET. I also thank Irby Lovette and Bronwyn Butcher from the Cornell Lab of Ornithology for their help during my visit to generate the genomic data. This project is funded by the BOU (small research grant awarded to Pablo D. Lavinia), the Richard Lounsbery Foundation, and the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) and the Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT) from Argentina.
References
Campagna, L., Kopuchian, C., Tubaro, P.L. & Lougheed, S.C. 2014. Secondary contact followed by gene flow between divergent mitochondrial lineages of a widespread Neotropical songbird (Zonotrichia capensis). Biological Journal of the Linnean Society 111(4): 863-868. VIEW
Campagna, L., Repenning, M., Silveira, L.F., Fontana, C.S., Tubaro, P.L. & Lovette, I.J. 2017. Repeated divergent selection on pigmentation genes in a rapid finch radiation. Science Advances 3(5): e1602404. VIEW
Funk, E.R. & Taylor, S.A. 2019. High-throughput sequencing is revealing genetic associations with avian plumage color. The Auk 136(4): ukz048. VIEW
Handford, P. 1983. Continental patterns of morphological variation in a South American sparrow. Evolution 37(5): 920–930. VIEW
Handford, P. 1985. Morphological relationships among subspecies of the rufous- collared sparrow, Zonotrichia capensis. Canadian Journal of Zoology 63(10): 2383–2388. VIEW
Küpper, C., Stocks, M., Risse, J.E., Dos Remedios, N., Farrell, L.L., McRae, S.B., Morgan, T.C., Karlionova, N., Pinchuk, P., Verkuil, Y.I., Kitaysky, A.S., Wingfield, J.C., Piersma, T., Zeng, K., Slate, J., Blaxter, M., Lank, D.B. & Burke, T. 2015. A supergene determines highly divergent male reproductive morphs in the ruff. Nature Genetics 48(1): 79–83. VIEW
Lougheed, S.C., Campagna, L., Dávila, J.A., Tubaro, P.L., Lijtmaer, D.A. & Handford, P. 2013. Continental phylogeography of an ecologically and morphologically diverse Neotropical songbird, Zonotrichia capensis. BMC Evolutionary Biology 13(1): 58. VIEW
Majidi, M., Hubbs, A.E. & Lichy, J.H. 1998. Activation of extracellular signal-regulated kinase 2 by a novel abl- binding protein, ST5. Journal of Biological Chemistry 273(26): 16608–16614. VIEW
Nottebohm, F. 1969. The Song of the Chingolo, Zonotrichia capensis, in Argentina: Description and Evaluation of a System of Dialects. The Condor 71(3): 299–315. VIEW
Rising, J. & Jaramillo, A. 2020. Rufous-collared Sparrow (Zonotrichia capensis). In J. del Hoyo, A. Elliott, J. Sargatal, D.A. Christie, & E. de Juana (Eds.) Handbook of the Birds of the World Alive. Lynx Edicions.
Tuttle, E.M., Bergland, A.O., Korody, M.L., Brewer, M.S., Newhouse, D.J., Minx, P., Stager, M., Betuel, A., Cheviron, Z.A., Warren, W.C., Gonser, R.A. & Balakrishnan, C.N. 2016. Divergence and functional degradation of a sex chromosome-like supergene. Current Biology 26(3): 344–350. VIEW
Vickery, A.I., Bruders, R., Kronenberg, Z., Mackey, E., Bohlender, R.J., Maclary, E.T., Maynez, R., Osborne, E.J., Johnson, K.P., Huff, C.D., Yandell, M. & Shapiro, M.D. 2018. Introgression of regulatory alleles and a missense coding mutation drive plumage pattern diversity in the rock pigeon. ELife 7. VIEW
Image credit
Featured image: Rufous-collared Sparrow Zonotrichia capensis australis from Santa Cruz, Argentina © Pablo D. Lavinia






