Report from a BOU-funded project
 The complex patterns of cold glacial and interglacial periods during the Pleistocene (2.59 MYA – 10,000 Y BP) caused repeated episodes of range expansions and contractions of populations, resulting in numerous episodes of vicariance and founder events. In Eurasia, southern peninsulas were refugia for species escaping the harsh climatic conditions, and in a few cases isolated populations diverged genetically and morphologically into new species in spite of occasional contact during warmer interglacial periods. This process of speciation could have been promoted by a combination of local adaptations and the establishment of behavioral and reproductive isolation mechanisms.
The complex patterns of cold glacial and interglacial periods during the Pleistocene (2.59 MYA – 10,000 Y BP) caused repeated episodes of range expansions and contractions of populations, resulting in numerous episodes of vicariance and founder events. In Eurasia, southern peninsulas were refugia for species escaping the harsh climatic conditions, and in a few cases isolated populations diverged genetically and morphologically into new species in spite of occasional contact during warmer interglacial periods. This process of speciation could have been promoted by a combination of local adaptations and the establishment of behavioral and reproductive isolation mechanisms.
A powerful way to assess the impact of these glacial periods and to recreate the process of speciation is through the comparison of the genomic variation between species pairs that shared a common ancestor that existed prior to these glacial periods. Here we will conduct whole mitochondria comparisons between two such sister taxa: the Spanish Imperial Eagle Aquila adalberti and the Eastern Imperial Eagle A. heliaca. Due to the very small population, divided into several breeding nuclei, and dependent on intensive management and conservation measures, the Spanish imperial eagle is listed as vulnerable under the IUCN red list of endangered species (BirdLife 2013a). Once abundant and widespread in the Iberian Peninsula, several threats such as poisoning, electrocution and insufficient food availability confined the population to the south-western quadrant of the Iberian Peninsula and left a population near extinction (Ferrer 2001). Thanks to years of strong efforts, the latest census showed around 500 breeding pairs in the wild, and a rising tendency since the last 15 years. Meanwhile, the Eastern Imperial Eagle population is estimated in 2,500-10,000 individuals across continental Eurasia and is not globally endangered. Although cautiously considered vulnerable by the UICN (BirdLife 2013b) due to small global population, presumed decline tendencies and threats of habitat loss and degradation, the situation for the Eastern Imperial Eagle is by far much better than for its Spanish relative. Improved knowledge of the complex evolutionary and demographic history of these species is key for the management and conservation of their populations.
We studied the Spanish Imperial Eagle population, and two Eastern Imperial Eagle populations, one in Hungary and one in Kazakhstan, representing the two geographical extremes of the species distribution, and we also included one individual sampled in Israel. The complete mitochondrial DNA was amplified and sequenced using next generation sequencing techniques for 11 Spanish and 11 Eastern Imperial Eagles. The complete mtDNA sequence of two Steppe Eagles A. nipalensis, the closest relative of imperial eagles, were also sequenced.
The recovered length of the mitochondrial genomes of Eastern Imperial Eagle, Spanish Imperial Eagle and Steppe Eagle were 17208 bp, 17207 bp and 17152 bp, respectively. This length is within the normal values for other species of birds and other organisms like mammals. Levels of diversity found are very low for the Spanish Imperial Eagle (Hd= 0,764+-0,083 SD, pi=0,00010, k=1,709), in agreement with previous studies using a small fraction of the mitochondria (Martinez-Cruz et al 2004, 2007). Higher levels of diversity are found in the Eastern Imperial Eagle (Hd= 0,1+-0,039 SD, pi=0,00063, k=10,909), although only four different haplotypes were found despite having sampled the two extremes of the species geographical range. Interestingly, only two individuals of Steppe Eagle, the nearest relative of the imperial eagles, and sampled in the same area both of them, showed a much higher level of diversity (Hd= 0,1+-0,5 SD, pi=0,00152, k=26). Apparently, as a common ancestor divided into two, one of the daughter species retained much less genetic diversity than the other along the evolutionary times, probably in parallel with a significantly lower population size.
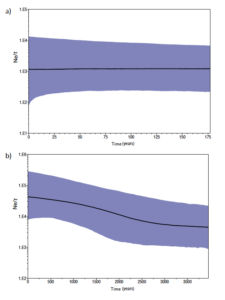 Figure 1. Evolution of the effective population size Ne through time estimated with BEAST for a) the Spanish Imperial Eagle, and b) the Eastern Imperial Eagle.
Figure 1. Evolution of the effective population size Ne through time estimated with BEAST for a) the Spanish Imperial Eagle, and b) the Eastern Imperial Eagle.
The study of the demographic evolution of the two imperial eagles when analysing the 13 coding genes of the mitochondria using the BEAST (Drummond et al 2012) method showed a completely different history between the two species (Figure 1). The Spanish Imperial Eagle shows a historically stable effective population size (Ne) of 103/t females during the last 100 years, where t is the generation time that could be approximated to 10 years. One hundred females is a low Ne, although higher than expected for a species that felt to a census size of 50 individuals in the last century (Ferrer 2001). Meanwhile, the demographic evolution of the Eastern Imperial Eagle shows an ascending trend in the last four thousand years, from 1,000 females Ne four thousand years ago up to 4,000 at present. Two points are important when interpreting these results. One is the forty times higher Eastern than Spanish imperial eagle effective population size, strengthen the idea of the weakness of the Spanish Imperial Eagle population. The second one is the difference in the demographic tendency, which interestingly for the Eastern imperial eagle shows an important increase in the last millennia, which are good news for the species.
Finally we analysed whether signs of adaptation could be found in the mitochondrial DNA of the imperial eagles species. We used already published mitochondrial sequences for other raptor species – Golden Eagle Aquila chrysaetos and four mountain hawk-eagle species Nisaetus nipalensis, Spizaetus nipalensis, S. alboniger and Spilornis cheela – to build a reference sequence against which to detect changes in the nucleotide composition of the thirteen mitochondrial genes in both imperial eagles that could alter the amino acid sequence and affect the encoded peptides. Although we detected twenty changes in the amino acid composition with respect to the reference sequence, sixteen of them fixed between the imperial eagle species, none of them seem to affect the function of the proteins they encode, indicating that no signals of adaptation are present in the mitochondria of imperial eagles.
The study of the whole mitochondria provides interesting insight into the demographic history and evolution of species. It is particularly interesting because it tells the story of the maternal lineage, allowing the comparison with the story for both sexes, through the study of the nuclear genome. Otherwise, it represents a single locus, and for this reason, is very limited. To follow up with our questions on demographic history and adaptation, we need to study the nuclear genome, as in deep as possible.
Funding
Begoña was awarded a BOU small ornithological grant of £1,05 for a project entitled ‘Maternal evolutionary history of imperial eagles Aquila heliaca and A. adalberti’ when she was a post-doctoral researcher at the Department of Integrative Biology, Doñana Biological Station (CSIC), Sevilla, Spain.
References
BirdLife International. 2013a. Spanish Imperial EagleAquila adalberti. The IUCN Red List of Threatened Species 2013: e.T22696042A48128913 VIEW.
BirdLife International. 2013b. Eastern Imperial Eagle Aquila heliaca. The IUCN Red List of Threatened Species 2013: e.T22696048A40763222. VIEW.
Drummond, A.J,, Suchard, M.A., Xie, D. & Rambaut, A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7 Molecular Biology and Evolution 29: 1969-1973 VIEW
Ferrer, M. 2001. The Spanish Imperial Eagle. Lynx Editions, Barcelona.
Martínez-Cruz, B., Godoy, J.A. & Negro, J.J. 2004. Population genetics after fragmentation: the case of the endangered Spanish imperial eagle (Aquila adalberti). Molecular Ecology 2004; 13: 2243-2255. VIEW
Martínez-Cruz, B., Godoy, J.A. & Negro, J.J. 2007. Population fragmentation leads to spatial and temporal genetic structure in the endangered Spanish imperial eagle. Molecular Ecology 2007; 16: 477-486. VIEW
Image credit
Top right: Eastern Imperial Eagle Aquila heliaca AngMoKio CC BY SA 2.5 Wikimedia Commons




