 LINKED PAPER
LINKED PAPER
Dietary niche overlap and resource partitioning among six steppe passerines of Central Spain using DNA metabarcoding. Zurdo, J., Barrero, A., da Silva, L. P., Bustillo-de la Rosa, D., Gómez-Catasús, J., Morales, M. B., Traba, J. & Mata, Vanessa A. 2023 Ibis. doi: 10.1111/ibi.13188 VIEW
Food niche partitioning is one of the main mechanisms through which many species limit interspecific competition, and therefore dietary niche overlap studies are of paramount importance for understanding community structure and the coexistence of ecologically similar species. In recent years, ornithologists and ecologists have taken advantage of DNA metabarcoding to characterize the diets of birds using faecal samples (Pompanon et al. 2012). This study represents the first application of faecal metabarcoding to assess interspecific trophic niche relationships in a shrub-steppe passerine community.
We collected 225 faecal samples of six passerine species that coexist in the southern ‘páramos’ (high-altitude, steppe-dominated plateaus) of Soria (central Spain): the endangered Dupont’s Lark (Chersophilus duponti), the Eurasian Skylark (Alauda arvensis), the Greater Short-toed Lark (Calandrella brachydactyla), the Tawny Pipit (Anthus campestris), the Northern Wheatear (Oenanthe oenanthe) and the Western Black-eared Wheatear (Oenanthe hispanica).

Figure 1 Male and female of one of the study species (Western Black-eared Wheatear Oenanthe hispanica), at the time of capture for the collection of faecal samples.
Samples were processed in the laboratory, where DNA was extracted and the eukaryotic mini-barcode miniB18S_81 designed by Cabodevilla et al. (2022) was used to amplify prey DNA, targeting the 18S rRNA gene. The final library was sequenced in an Illumina MiSeq NGS platform.
We identified 13 orders, 39 families, 27 genera and 14 species, all of them belonging to the phylum Arthropoda. Coleoptera was the most commonly identified order preyed upon across all bird samples, but regarding each bird species independently, the most frequent order differed, Orthoptera being the most common for the Eurasian Skylark, the Greater Short-toed Lark and the Western Black-eared Wheatear, and Coleoptera for Dupont’s Lark, the Tawny Pipit and the Northern Wheatear. Acrididae (grasshoppers), Julidae (millipedes), Tenebrionidae (darkling beetles) and Lycosidae (wolf spiders) were the most frequent preyed families.
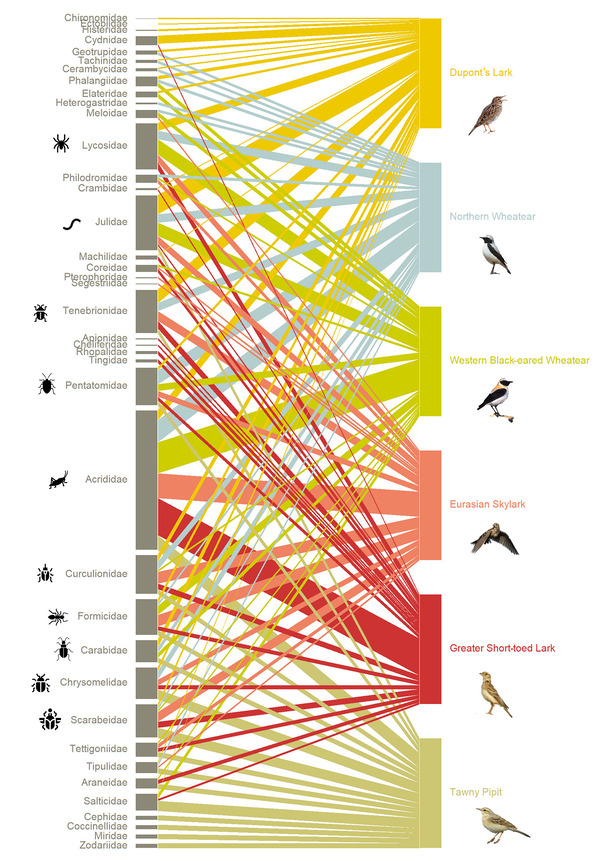
Figure 2. Network of the six passerine species’ dietary niche showing the frequency of occurrence of the prey families detected in each bird sample.
Niche overlap analysis returned a mean value of Pianka’s index of 0.74 for arthropod families, indicating a high overlap in the consumption of arthropod families. Lower niche overlap values were observed between Dupont’s Lark and the Greater Short-toed Lark and Tawny Pipit. We also found differences in prey composition between Dupont’s Lark and four other passerines (Eurasian Skylark, Greater Short-toed Lark, Western Black- eared Wheatear and Tawny Pipit) due to differences in the consumption of grasshoppers and millipedes.
The community-level overlap reported may represent the similarity of feeding strategies, as these shrub-steppe bird species forage mainly on the ground, by walking or running and picking up prey from the soil surface (de Juana et al. 2004). Also, it may reflect a peak period of arthropod abundance and availability, coinciding with these bird species’ breeding season, and indicate shared exploitation of abundant food resources.
The endangered Dupont’s Lark appeared to be the species with the most distinct dietary niche within the community and exhibited greater resource partitioning with the Tawny Pipit and, particularly, the Greater Short-toed Lark. These three species are passerines with very similar ecological traits and foraging habits that seem to be partitioning to some extent the available resources in the shrub-steppe environment to relax the effects of potential interspecific competition.
Further research with higher taxonomic resolution primer sets, including arthropod-specific COI markers, as well as covering the non-breeding season may describe species’ diets in greater detail and reveal trophic relationships that were undetected in this study.
References
Cabodevilla, X., Gómez-Moliner, B.J., Abad, N. & Madeira, M.J. 2022. Simultaneous analysis of the intestinal parasites and diet through eDNA metabarcoding. Integrative Zoology 1-15. VIEW
de Juana, E., Suárez, F., Ryan, P., Alström, P. & Donald, P. 2004. Family Alaudidae. In del Hoyo, J., Elliott, A. & Sargatal, J. (eds) Handbook of the Birds of the World, vol. 9: 496–601. Barcelona: Lynx Editions.
Pompanon, F., Deagle, B.E., Symondson, W.O.C., Brown, D.S., Jarman, S.N. & Taberlet, P. 2012. Who is eating what: Diet assessment using next generation sequencing. Molecular Ecology 21: 1931-1950. VIEW
Image credit
Top right: Dupont’s Lark Chersophilus duponti © Adrián Barrero.
If you want to write about your research in #theBOUblog, then please see here.





