LINKED PAPER
Highly contiguous genomes improve the understanding of avian olfactory receptor repertoires. Driver, R.J. & Balakrishnan, C.N. 2021 Integrative & Comparative Biology. doi: 10.1093/icb/icab150 VIEW
 One reason why birds capture our interest and imagination is that they share some of the same habits as humans. Unlike many mammals, most birds are diurnal and are often active at the same times of day as us. Stemming in part from their diurnal habits, birds, like humans, rely heavily on vision for foraging, competitive interactions, and attracting mates. As a result, many studies have described the incredible visual acuity of birds, including the morphological adaptations of raptor eyes and the ability of birds to detect a multitude of non-spectral colours (Mitkus et al. 2018, Stoddard et al. 2020). However, how birds perceive the world in other ways, such as through smell or olfaction, is not as well understood.
One reason why birds capture our interest and imagination is that they share some of the same habits as humans. Unlike many mammals, most birds are diurnal and are often active at the same times of day as us. Stemming in part from their diurnal habits, birds, like humans, rely heavily on vision for foraging, competitive interactions, and attracting mates. As a result, many studies have described the incredible visual acuity of birds, including the morphological adaptations of raptor eyes and the ability of birds to detect a multitude of non-spectral colours (Mitkus et al. 2018, Stoddard et al. 2020). However, how birds perceive the world in other ways, such as through smell or olfaction, is not as well understood.
Olfaction in birds suffers from some historical biases, including century-old behavioural experiments by John James Audubon as well as morphological studies that dissuaded the field of ornithology from giving birds credit for anything but a rudimentary sense of smell (Audubon 1826, Hill 1905). Debunking these convictions about bird smell began in the mid-20th century with more controlled and detailed behavioural experiments and morphological measurements of the olfactory bulb (Michelson 1959, Bang and Cobb 1968). Behavioural experiments in the past two decades have implicated bird olfaction in a variety of different behaviours including nest building, kin recognition, mate choice, and foraging (Wikelski 2021, see references in Driver & Balakrishnan 2021).
To perform these behaviours, birds then must have active olfactory receptors, the proteins that bind and detect compounds from the air to allow an animal to perceive smell. Each olfactory receptor may bind only one or several odorants, and so animals have many different kinds of olfactory receptors to detect and interpret a wide variety of smells. Mammals may have well over 1,000 olfactory receptors in their genome (Niimura et al. 2014), however, previous reports of bird olfactory receptor counts varied widely, with a few species showing hundreds of olfactory receptors, but the majority of birds show fewer than 50 (Khan et al. 2015).
Given that birds display behaviours associated with smell but often show very few olfactory receptors in their genomes, we performed our own investigation of bird olfactory receptor counts, published in Integrative & Comparative Biology. We noted that previous investigations into bird olfactory receptors did not use genomes along with long-read sequencing technology. Long-read sequencing technology allows for longer genomic regions to be sequenced at one time. For gene families with many members, such as olfactory receptors, two different genes that are very similar in sequence may be difficult to distinguish unless long-read sequencing is used. When long-read sequencing is implemented, similar genes can be more easily assigned as they reside on larger contigs, a series of overlapping DNA sequences, that are distinguishable from one another. Because many gene families are located in tandem along a contig, this leads to the potential characterization of many more genes that would otherwise be difficult to tell apart from one another.
We compared olfactory receptor repertoires that we could recover from genomes that used Sanger, short-read, and long-read sequencing methods. To our surprise, we found that not only do assemblies which use long-read sequencing recover additional olfactory receptors compared to short-read, but they recover many times more olfactory receptors, greatly broadening the number of olfactory receptors previously reported for these birds. For example, we found 27 olfactory receptors in the Anna’s Hummingbird (Calypte anna) short-read genome assembly, but 109 olfactory receptors in the long-read genome (Fig 1). This means that birds may be detecting a much wider variety of odorants that we previously thought possible.
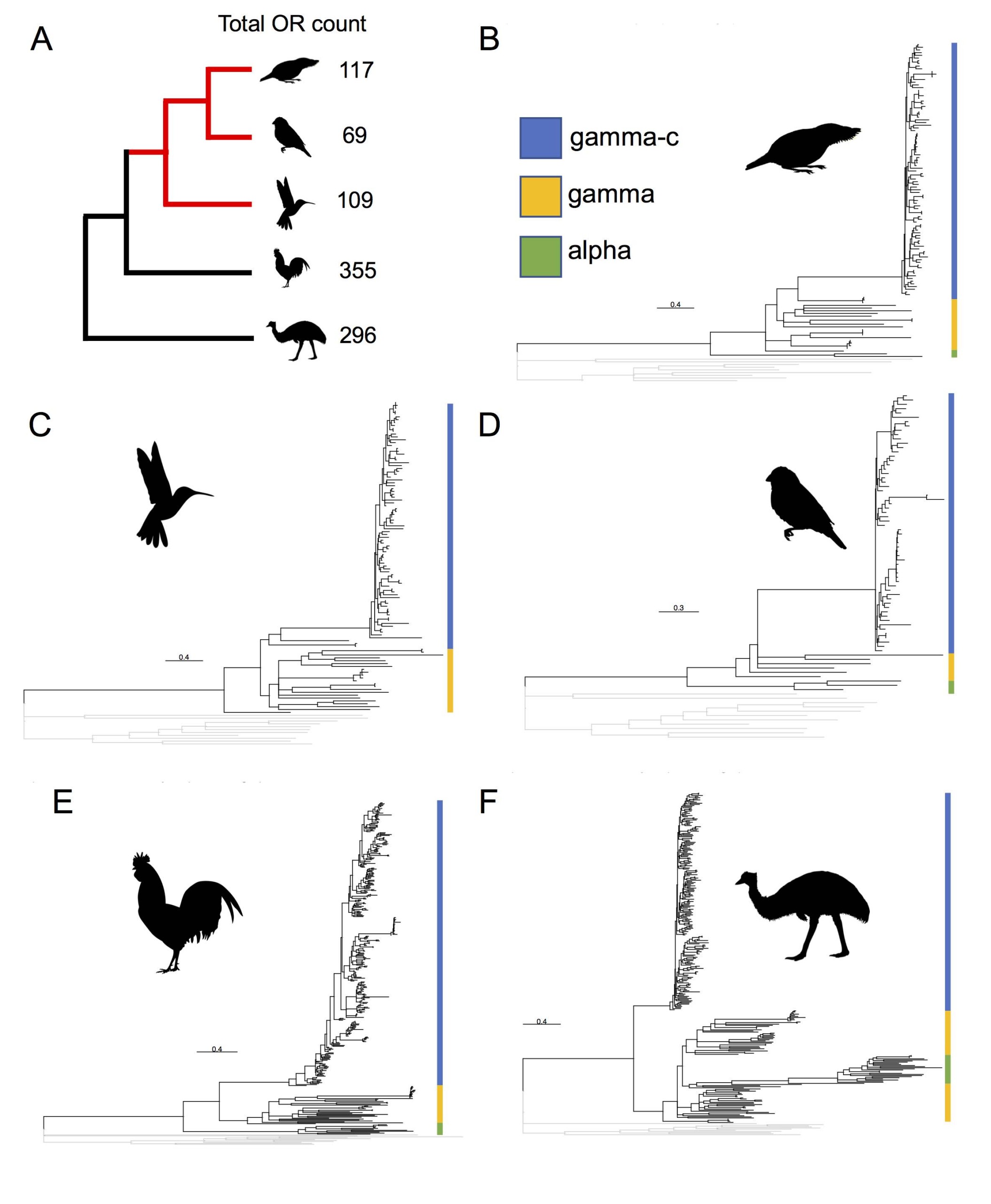
Figure 1 Phylogenetic reconstruction of olfactory receptor repertoires from long-read assemblies. (A) Topological phylogeny of species in this study, red branches indicate Neoaves species. Olfactory receptor counts from long-read genomes for each species are given. (B) The five species and assemblies shown are Golden-collared Manakin (Manacus vitellinus), (C) Anna’s Hummingbird (Calypte anna), (D) Zebra Finch (Taeniopygia guttata), (E) Red Junglefowl (Gallus gallus), (F) Emu (Dromaius novaehollandiae). The three OR subfamilies were assigned based on putative orthology to previously described bird ORs (Khan et al. 2015). Images not to scale. Bird images in E by Jim Bendon and F by Daniel Naish. Images E & F rasterized by Michael Keesey shared under CC BY 3.0. Image A is modified artwork by Kristen Orr.
Compared to short-read genomes, we detected increased numbers of olfactory receptors in all of the long-read genomes that we surveyed (Fig 1). We detected a particularly large increase in olfactory receptors belonging to a specific subfamily called the ‘gamma-c’, which are a group of olfactory receptors only found in birds, and also the most numerous kind of olfactory receptor in most bird species. We found this large number of gamma-c olfactory receptors in all the long-read bird genomes that we reported on, including the Emu (Dromaius novaehollandiae). Since this increased number of gamma-c olfactory receptors that are present in Emu, Chicken (Gallus gallus), and the Golden-collared Manakin (Manacus vitellinus), we show that the gamma-c olfactory receptor subfamily expansion is present across all major branches of the extant bird phylogeny. Therefore, the large number of gamma-c olfactory receptors was present in the ancestor of all modern birds, and it is likely that birds have relied on a sense of smell for tens of millions of years throughout their evolution.
Image credit
Top right: Golden-collared Manakin © Aaron Maizlish CC BY-NC 2.0 Flickr
References
Audubon, J.J. 1826. Account of the habits of the turkey buzzard, Vultur aura, particularly with the view of exploding the opinion generally entertained of its extraordinary power of smelling. The Edinburgh New Philosophical Journal 2: 172-184.
Bang, B.G. & Cobb, S. 1968. The size of the olfactory bulb in 108 species of birds. The Auk 85: 55-61. VIEW
Hill, A. 1905. Can birds smell? Nature 71: 18-19. VIEW
Khan, I., Yang, Z., Maldonado, E., Li, C., Zhang, G., Gilbert, M.T.P., Jarvis, E.D., O’Brien, S.J., Johnson, W.E. & Antunes, A. 2015. Olfactory receptor subgenomes linked with broad ecological adaptations in Sauropsida. Molecular Biology and Evolution 32: 2832-2843. VIEW
Michelsen, W.J. 1959. Procedure for studying olfactory discrimination in pigeons. Science 130: 630-631. VIEW
Mitkus, M., Potier, S., Marting, G., Duriez, O. & Kelber, A. 2018. Raptor vision. Oxford Research Encyclopedia of Neuroscience. VIEW
Niimura, Y., Matsui, A. & Touhara, K. 2014. Extreme expansion of the olfactory receptor gene repertoire in African elephants and evolutionary dynamics of orthologous gene groups in 13 placental mammals. Genome Research 24: 1485-1496. VIEW
Stoddard, M.C., Eyster, H.N., Hogan, B.G., Morris, D.H. Soucy, E.R. & Inouye, D.W. 2020. Wild hummingbirds discriminate nonspectral colors. Proceedings of the National Academy of Sciences 117: 15112-15122. VIEW
Wikelski, M., Quetting, M., Cheng, Y., Fiedler, W., Flack, A., Gagliardo, A., Salas, R., Zannoni, N. & Williams, J. 2021. Smell of green leaf volatiles attracts white storks to freshly cut meadows. Scientific Reports 11: 12912. VIEW





