LINKED PAPER
Genetic drift and bottleneck do not influence diversity in Toll-like receptor genes at a small spatial scale in a Himalayan passerine. Nandakumar, M. & Ishtiaq, F. 2020 Ecology and Evolution. DOI: 10.1002/ece3.6855 VIEW
 How do birds handle a high pathogenic environment? Are they equipped to cope if a new pathogen is introduced in their environment? To address these questions, the exhaustive way is to observe and track various bird populations in different pathogenic environments, across generations and quantify their immune response. But a simpler way would be to look at genetic sequences in congeneric species and compare their genetic diversity. Estimates of genetic diversity, i.e. studying variations in gene sequences, are useful metrics that provide insights into the evolution of a population. Populations with higher genetic diversity are known to adapt better to a changing environment and therefore have increased fitness. Within the context of pathogens, the most informative gene candidates to quantify genetic diversity would then be the genes of the immune system. It is expected that in a high pathogenic environment, there is an accompanied increase in genetic diversity of the immune genes. Populations survive better since they can recognise and respond to diverse pathogens.
How do birds handle a high pathogenic environment? Are they equipped to cope if a new pathogen is introduced in their environment? To address these questions, the exhaustive way is to observe and track various bird populations in different pathogenic environments, across generations and quantify their immune response. But a simpler way would be to look at genetic sequences in congeneric species and compare their genetic diversity. Estimates of genetic diversity, i.e. studying variations in gene sequences, are useful metrics that provide insights into the evolution of a population. Populations with higher genetic diversity are known to adapt better to a changing environment and therefore have increased fitness. Within the context of pathogens, the most informative gene candidates to quantify genetic diversity would then be the genes of the immune system. It is expected that in a high pathogenic environment, there is an accompanied increase in genetic diversity of the immune genes. Populations survive better since they can recognise and respond to diverse pathogens.
However, there are several confounding factors that influence genetic diversity that must also be ascertained. The extent of genetic diversity is in fact determined by a combination of population history (population expansion vs contraction), random chance (genetic drift), and selection pressure based on the genomic context (functional importance of the genomic region). While the effects of genetic drift and population history on variation is uniform across all regions of the genome, in functionally important genes selection pressure from the environment (such as pathogen pressure on immune genes) mainly influences diversity. We can understand which factor is dominant by comparing diversity in a functionally important genomic region (“adaptive” regions or loci) with a control region that is non-functional (“neutral” regions or loci). This is the crux of our study. We used genetic data from a host-parasite system of two Himalayan passerine species found along different elevations and their blood parasites (Plasmodium, Haemoproteus and Leucocytozoon), to answer questions related to pathogen-mediated selection pressure shaping immune genetic diversity. In our system, elevation served as a pathogen gradient, with fewer blood parasites found as you move up the elevation (Ishtiaq and Barve, 2018).
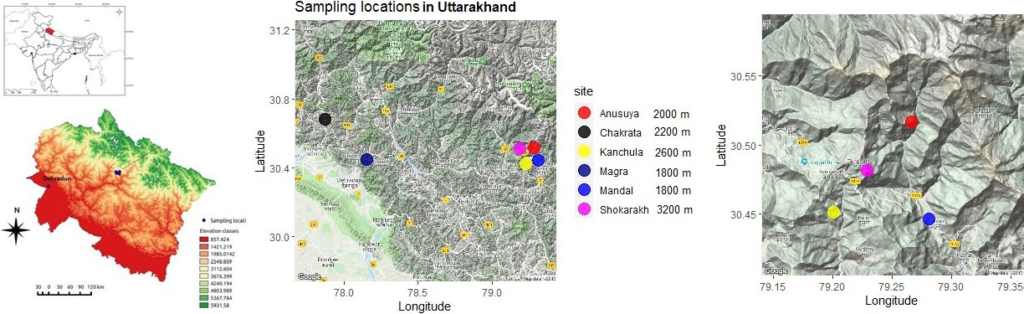
We sampled Black-throated Tit (BTT), Aegithalos concinnus (recently split as Aegithalos iredalei in India), across five sites at different elevations (1800–2600 m.a.s.l.). The BTTs are sedentary passerines found across different regions of Asia, including India and China. For each BTT sample, we quantified diversity in a set of microsatellites (“neutral loci”) and toll-like receptors (TLRs; “adaptive loci”). TLRs are a set of immune genes that act as pathogen sensors, forming the first line of immune defence. Using the microsatellite data, we first established population information—the BTT populations disperse a lot between sampling locations (range of 1.5–150 km) and elevations, and have no population genetic structure. We then compared the extent of genetic diversity between microsatellites and TLRs within each BTT population. We found a pattern such that at every elevation, there is significantly higher diversity in the TLR genes than the microsatellites. This tells us that there is selection pressure, possibly from pathogens, acting on TLRs that increases genetic diversity. However, we don’t find any differences in TLR genetic diversity between elevations.
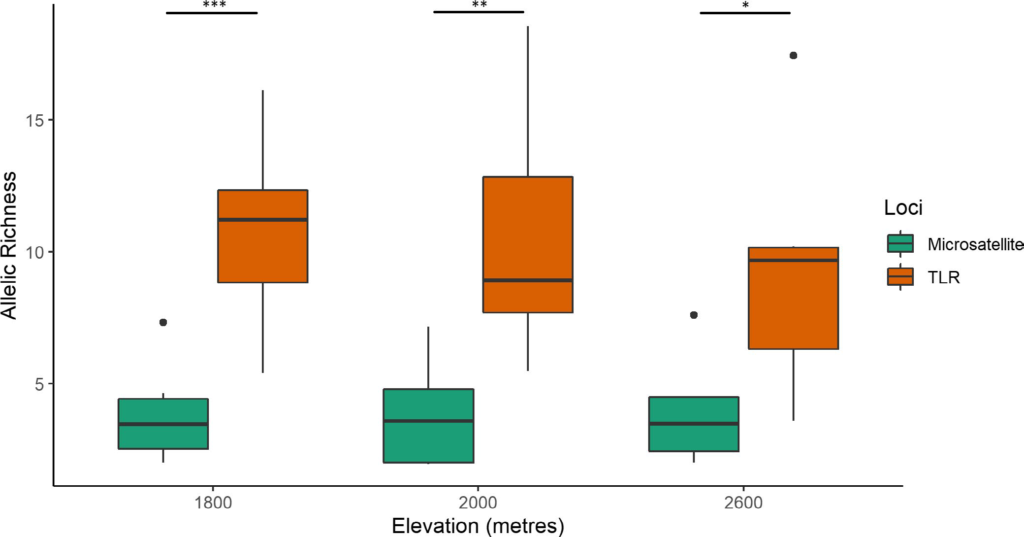 Figure 2 Comparison of genetic diversity (as allelic richness) between microsatellites and TLRs in different BTT populations.
Figure 2 Comparison of genetic diversity (as allelic richness) between microsatellites and TLRs in different BTT populations.
To investigate if demographic events caused the low microsatellite diversity, we compared our microsatellite data with another sister population in China (Dai et al., 2017). The BTT populations had indeed undergone a past population bottleneck event which led to a dramatic reduction in population, and consequently, genetic diversity. These findings are in contrast with other studies on bottlenecked populations of endangered species, where low genetic diversity in both adaptive and neutral loci was the norm.
Is selection pressure exerted by pathogens determined by an elevation gradient? To answer this, we quantified genetic diversity in TLRs of a high-elevation sister species, Aegithalos niveogularis (White-throated Tit; WTT). The WTTs are sedentary passerines found only at a high elevation of 3200 m.a.s.l.
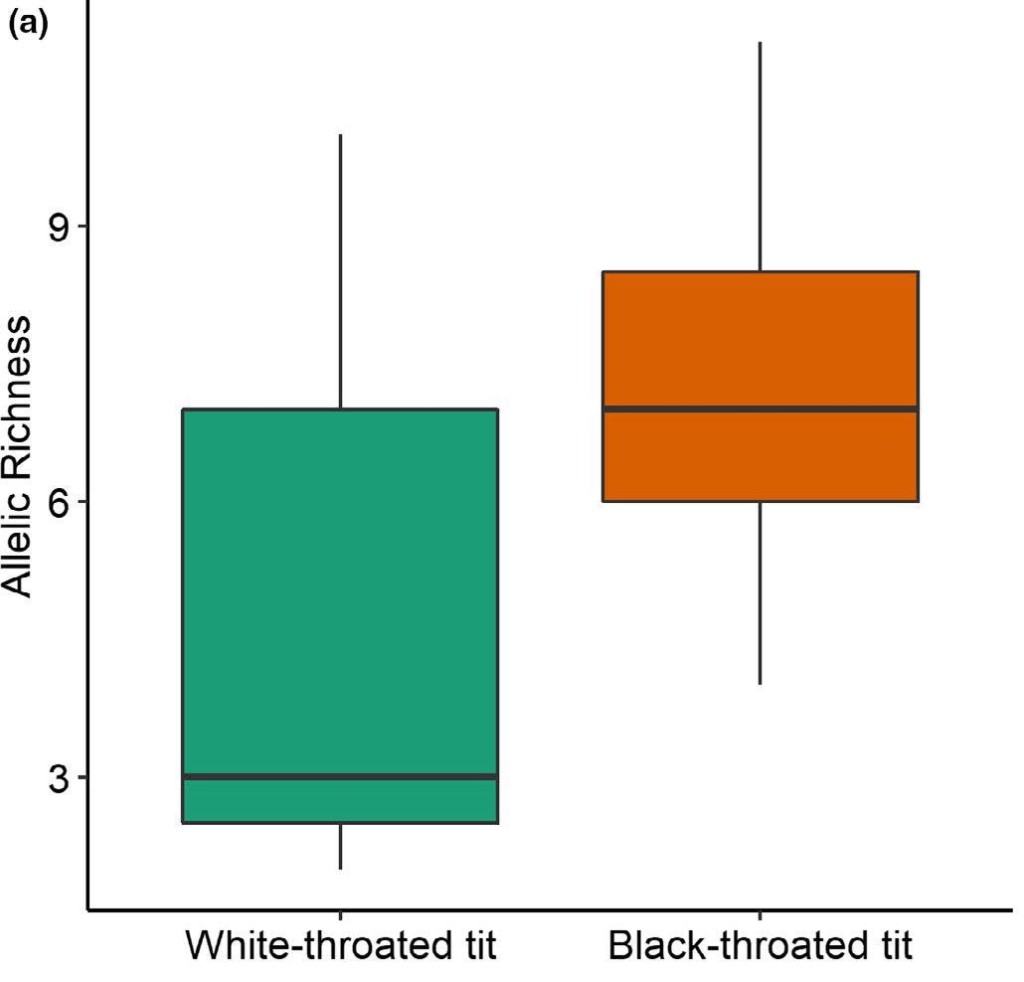 Figure 3 Comparison of genetic diversity (as allelic richness) between low- to mid-elevation BTTs and high-elevation WTTs.
Figure 3 Comparison of genetic diversity (as allelic richness) between low- to mid-elevation BTTs and high-elevation WTTs.
Contrary to our expectations, we found no significant differences in TLR genetic diversity between the BTTs and WTTs, probably due to the low sample size of the latter. In fact, the targets of selection loci in two TLR genes were found to be the same in BTTs and WTTs. Due to sampling constraints and the low WTT sample size, we did not find an effect of parasite prevalence on TLR diversity. However, in general there is a high prevalence of Leucocytozoon parasites in high-elevation birds, which could reflect the similar pathogenic environment experienced by the two passerines. As a follow up, it would be great to look into this using a large sample size and parasite lineage-specific relationship across high elevation species.
Nonetheless, the results from this study show that reduction in population size could lead to reduction in neutral genetic diversity but high immune gene diversity that helps in the revival of bottlenecked populations.
References
Ishtiaq, F. & Barve, S. 2018. Do avian blood parasites influence hypoxia physiology in a high elevation environment? BMC Ecology 18: 15. VIEW
Dai, C., Hao, Y., He, Y. & Lei, F. 2017. The absence of reproductive isolation between non-sister and deeply diverged mitochondrial lineages of the black-throated tit (Aegithalos concinnus) revealed by a multilocus genetic analysis in a contact zone. BMC Evolutionary Biology 17: 266. VIEW
Image credit
Top right: Black-throated Tit Aegithalos concinnus | Robert tdc, CC BY-SA 2.0, via Wikimedia Commons




